World and internet is full of textual information. We search for information using textual queries, we read websites, books, e-mails. All those are strings from the point of view of computer science. To make sense of all that information and make search efficient, search engines use many string algorithms. Moreover, the emerging field of personalized medicine uses many search algorithms to find disease-causing mutations in the human genome. In this online course you will learn key pattern matching concepts: tries, suffix trees, suffix arrays and even the Burrows-Wheeler transform.

Algorithms on Strings

Algorithms on Strings
This course is part of Data Structures and Algorithms Specialization



Instructors: Neil Rhodes
97,250 already enrolled
Included with
1,090 reviews
Skills you'll gain
Details to know

Add to your LinkedIn profile
4 assignments
See how employees at top companies are mastering in-demand skills

Build your subject-matter expertise
- Learn new concepts from industry experts
- Gain a foundational understanding of a subject or tool
- Develop job-relevant skills with hands-on projects
- Earn a shareable career certificate

There are 4 modules in this course
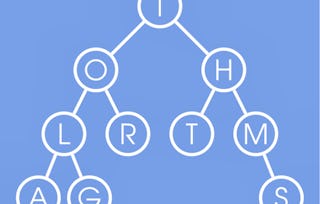
How would you search for a longest repeat in a string in LINEAR time? In 1973, Peter Weiner came up with a surprising solution that was based on suffix trees, the key data structure in pattern matching. Computer scientists were so impressed with his algorithm that they called it the Algorithm of the Year. In this lesson, we will explore some key ideas for pattern matching that will - through a series of trials and errors - bring us to suffix trees.
What's included
6 videos5 readings1 assignment1 programming assignment
Although EXACT pattern matching with suffix trees is fast, it is not clear how to use suffix trees for APPROXIMATE pattern matching. In 1994, Michael Burrows and David Wheeler invented an ingenious algorithm for text compression that is now known as Burrows-Wheeler Transform. They knew nothing about genomics, and they could not have imagined that 15 years later their algorithm will become the workhorse of biologists searching for genomic mutations. But what text compression has to do with pattern matching??? In this lesson you will learn that the fate of an algorithm is often hard to predict – its applications may appear in a field that has nothing to do with the original plan of its inventors.
What's included
5 videos4 readings1 assignment1 programming assignment
Congratulations, you have now learned the key pattern matching concepts: tries, suffix trees, suffix arrays and even the Burrows-Wheeler transform! However, some of the results Pavel mentioned remain mysterious: e.g., how can we perform exact pattern matching in O(|Text|) time rather than in O(|Text|*|Pattern|) time as in the naïve brute force algorithm? How can it be that matching a 1000-nucleotide pattern against the human genome is nearly as fast as matching a 3-nucleotide pattern??? Also, even though Pavel showed how to quickly construct the suffix array given the suffix tree, he has not revealed the magic behind the fast algorithms for the suffix tree construction!In this module, Miсhael will address some algorithmic challenges that Pavel tried to hide from you :) such as the Knuth-Morris-Pratt algorithm for exact pattern matching and more efficient algorithms for suffix tree and suffix array construction.
What's included
8 videos2 readings1 assignment
In this module we continue studying algorithmic challenges of the string algorithms. You will learn an O(n log n) algorithm for suffix array construction and a linear time algorithm for construction of suffix tree from a suffix array. You will also implement these algorithms and the Knuth-Morris-Pratt algorithm in the last Programming Assignment in this course.
What's included
16 videos3 readings1 assignment1 programming assignment
Earn a career certificate
Add this credential to your LinkedIn profile, resume, or CV. Share it on social media and in your performance review.
Instructors

Offered by
Explore more from Algorithms
 Status: Free Trial
Status: Free TrialJohns Hopkins University
 Status: Free Trial
Status: Free TrialGoogle
 Status: Free Trial
Status: Free TrialUniversity of California San Diego
 Status: Free
Status: FreePrinceton University
Why people choose Coursera for their career

Felipe M.

Jennifer J.

Larry W.

Chaitanya A.
Learner reviews
- 5 stars
67.43%
- 4 stars
21%
- 3 stars
7.52%
- 2 stars
2.29%
- 1 star
1.74%
Showing 3 of 1090
Reviewed on Jun 5, 2017
Unfortunately the forums go inactive after the first few iterations of the course. One can still learn by doing the programming assignments
Reviewed on Sep 8, 2016
Still stuck at Problem 5 of Week 1, had to left this problem undone.Wondering if the mentors or professors can send those who completed the course the standard/reference solution/program via Email???
Reviewed on Jan 24, 2017
Initially the accent was a little bit hard to understand, but after few minutes everything become crystal clear. Extremely useful course content.

Open new doors with Coursera Plus
Unlimited access to 10,000+ world-class courses, hands-on projects, and job-ready certificate programs - all included in your subscription
Advance your career with an online degree
Earn a degree from world-class universities - 100% online
Join over 3,400 global companies that choose Coursera for Business
Upskill your employees to excel in the digital economy
Frequently asked questions
To access the course materials, assignments and to earn a Certificate, you will need to purchase the Certificate experience when you enroll in a course. You can try a Free Trial instead, or apply for Financial Aid. The course may offer 'Full Course, No Certificate' instead. This option lets you see all course materials, submit required assessments, and get a final grade. This also means that you will not be able to purchase a Certificate experience.
When you enroll in the course, you get access to all of the courses in the Specialization, and you earn a certificate when you complete the work. Your electronic Certificate will be added to your Accomplishments page - from there, you can print your Certificate or add it to your LinkedIn profile.
Yes. In select learning programs, you can apply for financial aid or a scholarship if you can’t afford the enrollment fee. If fin aid or scholarship is available for your learning program selection, you’ll find a link to apply on the description page.
More questions
Financial aid available,



